$173 Million Study into Human Microbiome
The highly respected and influential journal, Nature yesterday published the results of a vast study into the microbes (bacteria, fungi and viruses) found in and upon the human body.
Within this article:
The Human Microbiome Project analysed an impressive 4,788 specimens from 242 adults. The project found enormous variations in microbial populations from person to person, even though all participants were deemed to be healthy. Although the study has not gone so far as to suggest which bacteria cause or prevent diseases, this large-scale, well-funded and widely published research is exciting as it demonstrates the growing medical focus on human bacteria in the gut and the rest of the body.
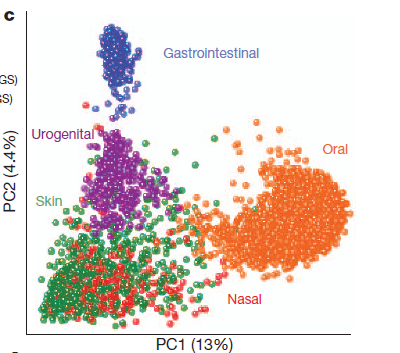
The article published in Nature acknowledged that previous studies had linked a low diversity of microbes to obesity and inflammatory bowel disease, and high diversity in the vagina to bacterial vaginosis. This study found vaginal sites to harbour particularly simple communities of microorganisms across different subjects in the study, featuring a high number of bacteria from the Lactobacillus genus. The microbes studied in oral & stool communities were found to be particularly diverse from participant to participant.
For those participants whose microbiomes were measured over time, their microbial populations were indeed found to vary with time, but not nearly to the extent that microbiomes varied from individual to individual.
The study is, rightly so, a hot topic in the health & medicinal press at the moment - the New York Times yesterday published a piece entitled In Good Health? Thank you 100 Trillion Bacteria with a particular focus on the microorganisms deemed to be beneficial - i.e. probiotics.
Methods
Microbial samples were taken from female participants at 18 body habitats, and from men at 15 (the same, but excluding 3 vaginal sites). Collection points included diverse areas such as the gums, cheek, throat, tongue, nostrils, inner elbows, the posterior fornix in women, and stool samples for the lower intestinal tract. 131 participants had additional samples taken at a later date in order to evaluate stability of individual microbiomes over time.
Possible Implications
Scientists do hope that this Human Microbiome Project will pave the way for further research in the area. Perhaps in the future analysing a person's microbes might serve to tell doctors and scientists who may have, or be at risk of, which disease. The study points towards further interest in modifying a person's microbiome (the name given to the collection of all the bacteria, fungi, and viruses carried by a person).
Either way, its all very exciting for the world of microbes. Professor Ger Rijkers, PhD, from the University Medical Center in Utrecht, in the Netherlands, said 'The data looks great and will most certainly advance further research into probiotics and prebiotics' (see Probiotics Learning Lab for more information).
Let's hope so!
Find out more about The Human Microbiome Project.
References
- Huttenhower, C., Gevers, D., Knight, R. et al. (2012). Structure, function and diversity of the healthy human microbiome. Nature, 486: 207–214. https://doi.org/10.1038/nature11234
- Herper, M. (2012). Government Unveils First Map Of All The Germs In The Human Body. [online] Forbes. Available at: <http://www.forbes.com/sites/matthewherper/2012/06/13/government-unveils-first-map-of-all-the-germs-in-the-human-body/> [Accessed 12 July 2012].
- Kolata, G. (2012). In Good Health? Thank Your 100 Trillion Bacteria. [online] Available at: <http://www.nytimes.com/2012/06/14/health/human-microbiome-project-decodes-our-100-trillion-good-bacteria.html?pagewanted=all> [Accessed 12 July 2012].


